Bioinformatics & Metabolomics Unit
The Bioinformatics & Metabolomics Unit is a multidisciplinary research and support platform housed within the Center for Immunometabolism (CIM) at the University of Pittsburgh. The unit is dedicated to advancing the analysis, interpretation, and visualization of high-dimensional biological data generated across a wide range of experimental platforms and clinical cohorts.
Leveraging expertise in bioinformatics, metabolomics, and multi-omics data integration, the unit plays a pivotal role in supporting translational and systems-level research. Our team collaborates closely with investigators in immunology, cardiometabolism, oncology, endocrinology, and related fields to extract meaningful biological insights from complex datasets, ultimately driving discoveries that inform both basic science and clinical applications.
Key capabilities include, but are not limited to, the analysis of genomic, transcriptomic, proteomic, metabolomic, and single-cell RNA sequencing data, as well as spatial transcriptomics. The unit also leverages machine learning and artificial intelligence techniques to mine large-scale datasets, enabling the identification of predictive biomarkers, elucidation of disease mechanisms, and discovery of novel therapeutic targets.
Mission
To provide researchers with state-of-the-art tools, customized assays, and expert consultation to dissect cellular metabolic pathways, enabling the discovery of novel therapeutic targets for metabolic diseases.
- Advance our understanding of complex biological systems
- Support the discovery of robust biomarkers and therapeutic targets
- Enable personalized and precision medicine approaches
- Facilitate early disease detection, patient stratification, and treatment optimization
The unit serves as both a collaborative hub and an innovation engine, accelerating the pace of research from bench to bedside through rigorous data-driven science.
Key Focus Areas
Bioinformatics
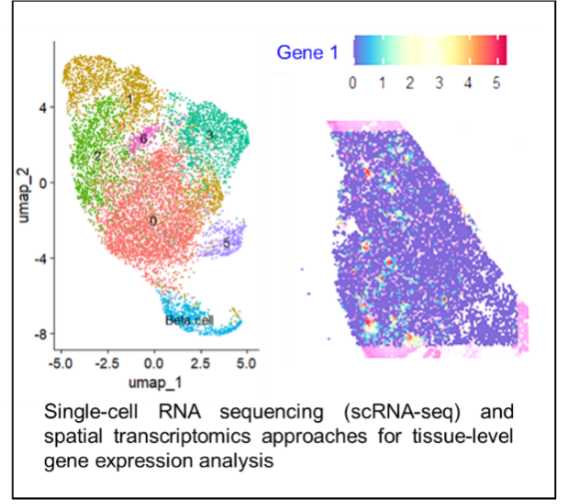
Comprehensive computational analysis of genomic, transcriptomic, proteomic, metabolomic, and single-cell sequencing data, with a focus on biomarker discovery, disease stratification, and unraveling disease-associated molecular mechanisms. The unit also provides support for spatial transcriptomics, enabling the integration of gene expression data with tissue architecture for deeper biological insights.
Machine Learning & Artificial Intelligence
Application of advanced data science and machine learning techniques to extract meaningful patterns from large-scale omics datasets and cohort-based clinical data.
Human cohort analysis
Integration and analysis of clinical, molecular, and phenotypic data from human cohorts to identify disease risk factors, progression markers, and patient subgroups. This includes support for longitudinal studies and population-based research.
Highlighted Work-Related References
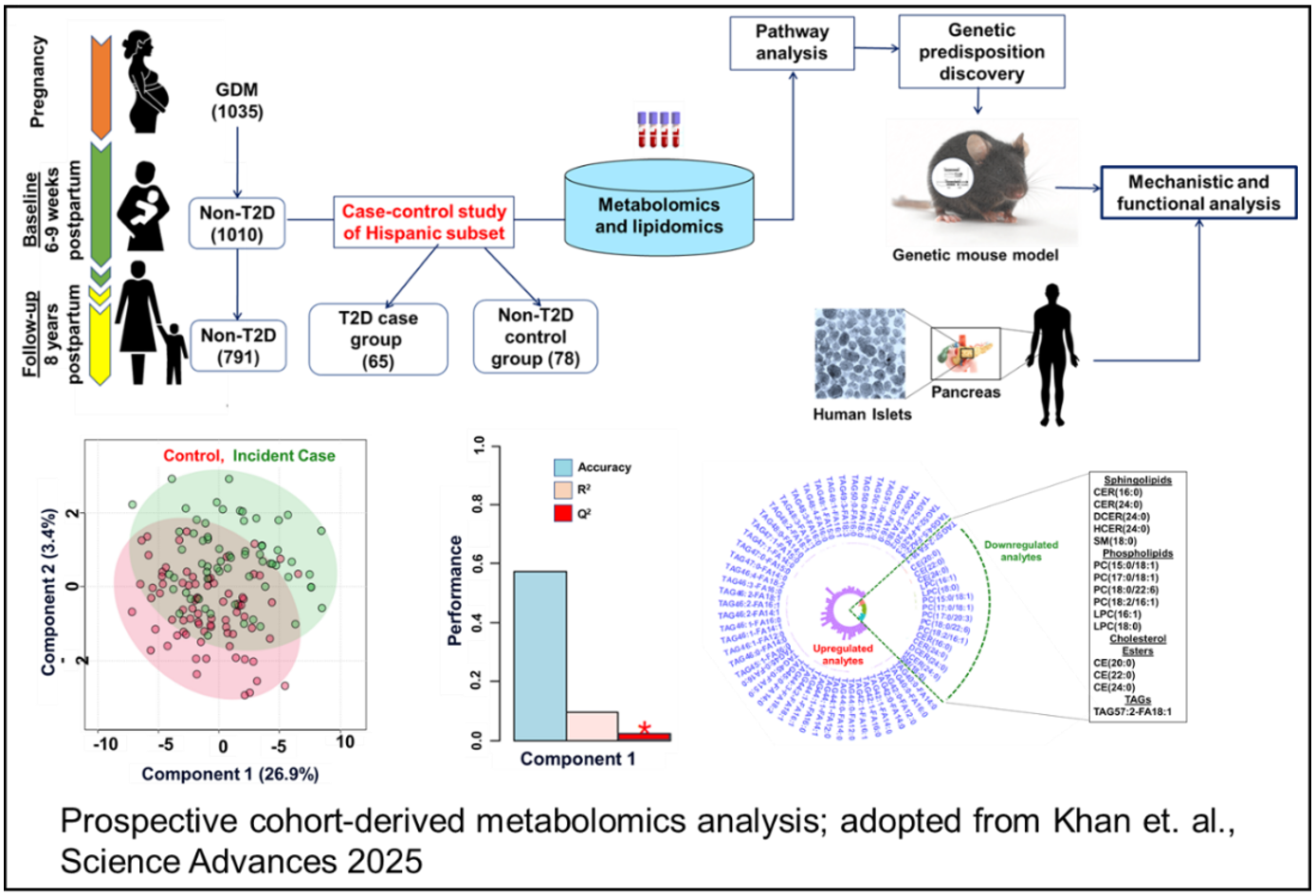
Khan, S.R.* et al. (2025). Reduced circulating sphingolipids and CERS2 activity are linked to T2D risk and impaired insulin secretion. Science Advances, 11, eadr1725. (Corresponding author)
Khan, S.R.*, et al. (2025). Early postpartum metabolic heterogeneity among women who progressed to type 2 diabetes after gestational diabetes: A prospective cohort. Diabetes Metabolism Research and Reviews, 41, e70027. (Corresponding author)
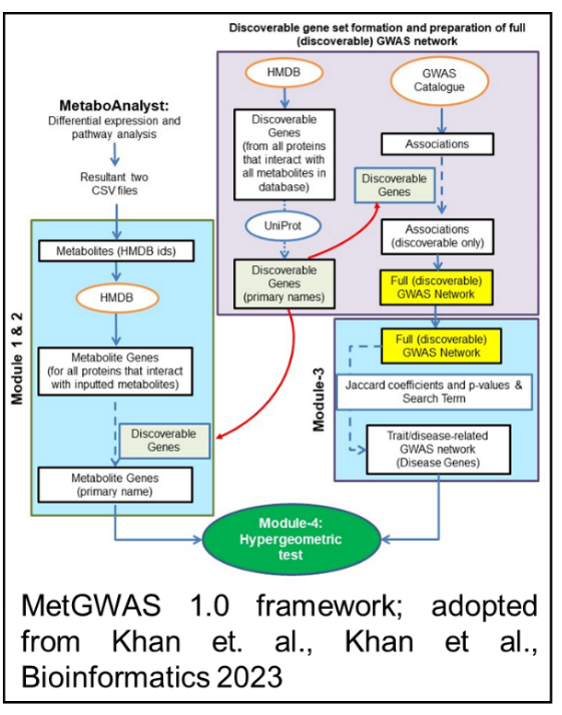
*Khan, S.R.*, et al. (2023). metGWAS 1.0: Network-driven over-representation analysis between independent metabolomic and genome-wide association studies identifies gene–metabolite relationships to phenotype. Bioinformatics, 39(9). (Corresponding author)
Khan, S.R.*, et al. (2021). Integration of AI and traditional medicine in drug discovery. Drug Discovery Today. (Corresponding author)
Khan, S.R., et al. (2020). Diminished sphingolipid metabolism—a hallmark of future type 2 diabetes pathogenesis—is linked to pancreatic beta-cell dysfunction. iScience, 23(10).
Manialawy, Y., Khan, S.R*., et al. (2020). The magnesium transporter NIPAL1 is a novel pancreatic islet–expressed protein that impacts insulin secretion. Journal of Biological Chemistry, 295(29), 9879–9892. (Corresponding author)
Khan, S.R*., et al. (2019). Unbiased data analytic strategies to improve biomarker discovery in precision medicine. Drug Discovery Today, 24(9), 1735–1748. (Corresponding author)
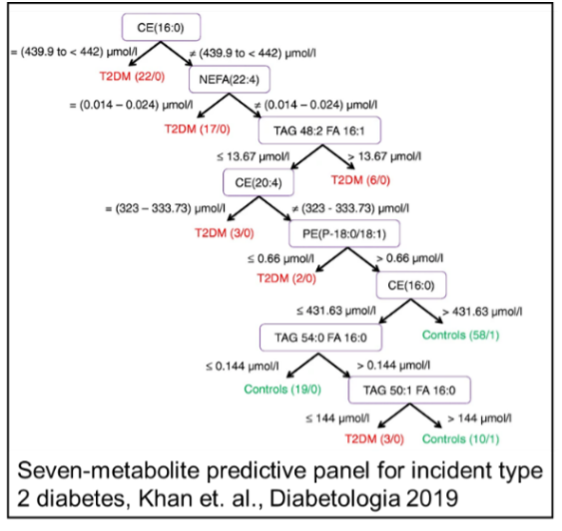
Khan, S.R., et al. (2019). The discovery of novel predictive biomarkers and early-stage pathophysiology for the transition from gestational diabetes mellitus to type 2 diabetes mellitus. Diabetologia, 62(4), 687–703.