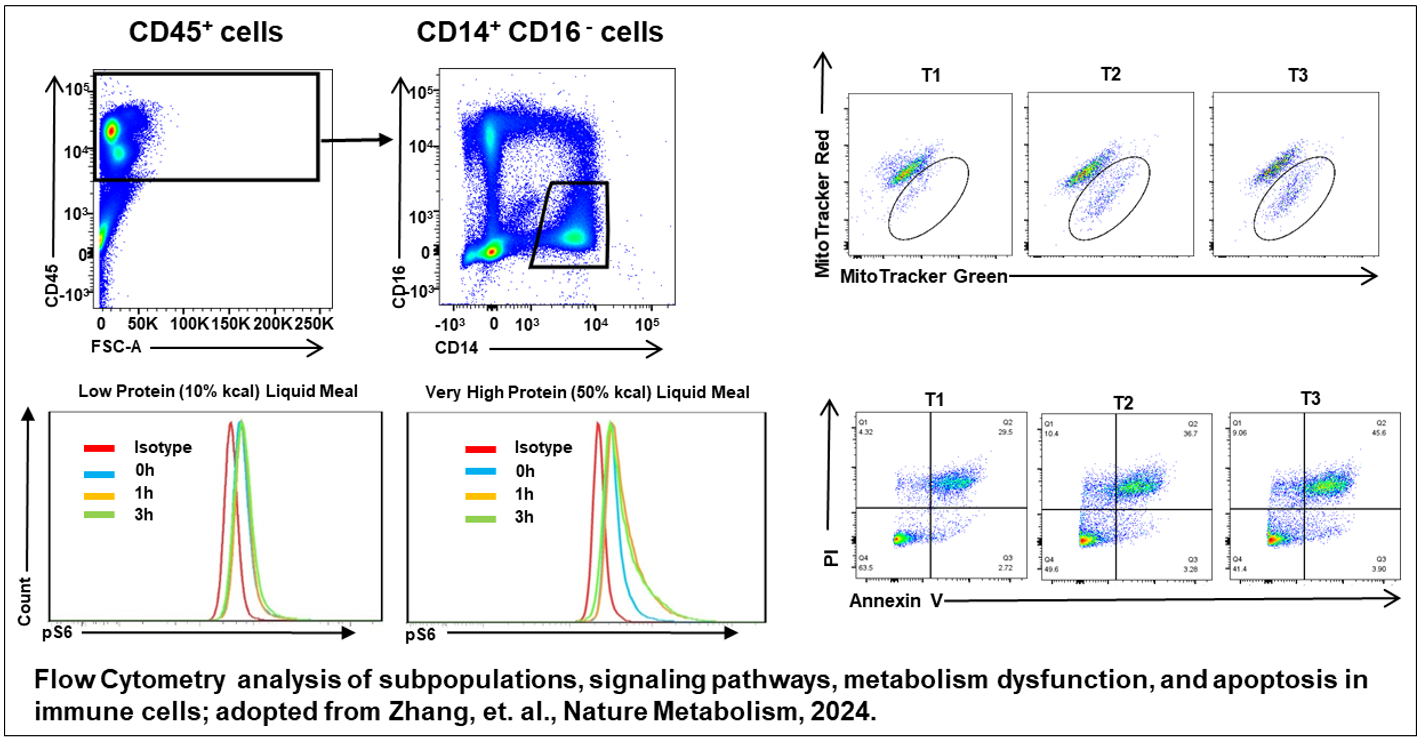
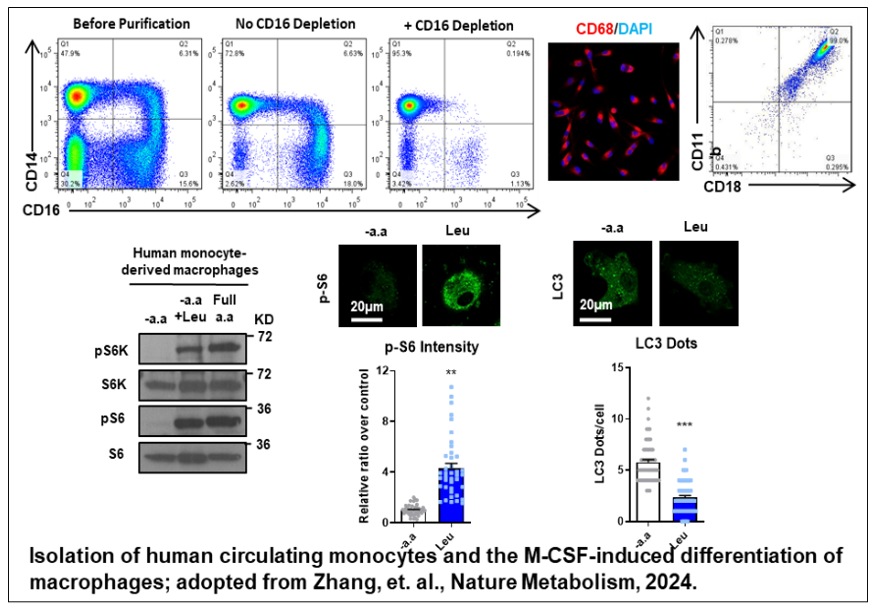
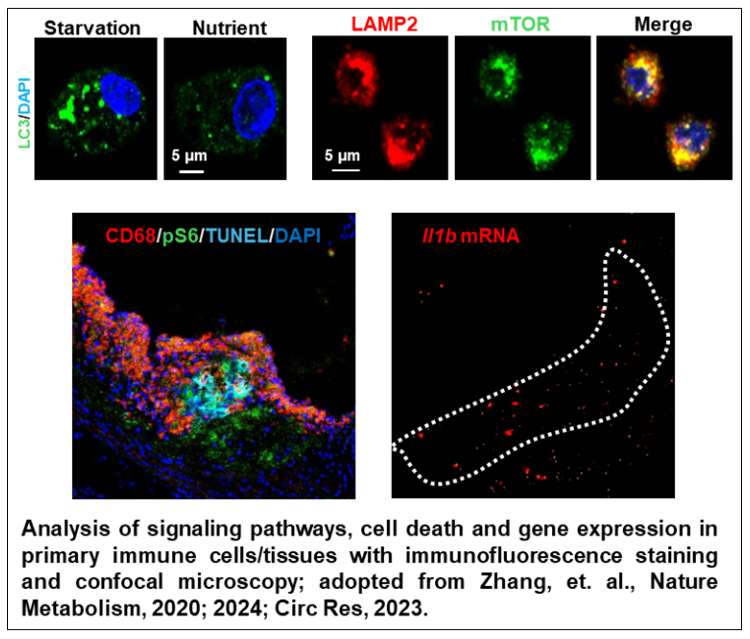
Immune Phenotyping Unit
Key capabilities include, but are not limited to, the analysis of immune cell subpopulations, intracellular cytokine expression, signaling pathways, metabolic dysfunction, and cell death using flow cytometry. The Unit also provides cell sorting services and guidance on in vitro culture and differentiation of isolated cells. In addition, the Unit assists researchers with the rapid examination of signaling pathways, metabolic dysfunction, and gene expression using immunofluorescence staining and confocal microscopy.
Mission
By providing cutting-edge analytical support, the Unit aims to:
- Advance the understanding of immunometabolism in disease pathogenesis
- Characterize unique subsets of immune cells and signaling pathways involved in disease development
- Support the development of novel and precision therapeutic approaches targeting immune cells
Key Focus Areas
Nutrient sensing and metabolism in immune cells
Through collaboration with the CIM Bioinformatics & Metabolomics Unit, distinctive nutrient-sensing and metabolism-related genes and signaling pathways in immune cell subpopulations are identified via comprehensive computational analyses of genomic, transcriptomic, proteomic, metabolomic, and single-cell sequencing data. Flow cytometry and confocal microscopy are then employed to determine the roles of these target genes and pathways in disease pathogenesis.
Human biopsy and clinical studies
To develop personalized and precision medicine approaches, metabolic disease-related human biopsies are being collected for the construction of the CIM Biobank. Findings from animal models will be validated in these human samples using flow cytometry and other techniques. In addition, the Unit supports researchers in developing plans and establishing collaborations for clinical trials.
Highlighted Work-Related References
Zhang XY, et al. Identification of a leucine-mediated threshold effect governing macrophage mTOR signaling and cardiovascular risk. Nat. Metab; 2024; 6(2):359-377.
Zhang XY, et al. Loss of Macrophage mTORC2 Drives Atherosclerosis via FoxO1 and IL-1β Signaling. Circ Res; 2023; 133(3):200-219.
Zhang XY, et al. Use of Acidic Nanoparticles to Rescue Macrophage Lysosomal Dysfunction in Atherosclerosis. Autophagy; 2023; 19(3):886-903.
Zhang XY, Stitham J, Rodriguez-Velez A, Jeong SJ, Park A, Yeh YS, Kapoor D, Mittendorfer D, Razani, B. Evaluation of mTORC1 signaling in mouse atherosclerotic macrophages by flow cytometry and immunofluorescence. STAR protocols; 2022; 3 (4), 101665.
Zhang XY, et al. High-protein diets increase cardiovascular risk by activating macrophage mTOR to suppress mitophagy. Nat Metab; 2020; 2(1):110-25.
Evans TD, Zhang XY, et al. Functional Characterization of LIPA (Lysosomal Acid Lipase) Variants Associated With Coronary Artery Disease. Arterioscler Thromb Vasc Biol; 2019; 39(12):2480-91.
Evans TD, Zhang XY, et al. TFEB drives PGC-1α expression in adipocytes to protect against diet-induced metabolic dysfunction. Sci Signal; 2019; 12(606): eaau2281.
Sergin I, Evans TD, Zhang XY, et al. Exploiting macrophage autophagy-lysosomal biogenesis as a therapy for atherosclerosis. Nat Commun; 2017; 8:15750-69.